2 回答
TA贡献1777条经验 获得超10个赞
在完全周期性的情况下,我会做这样的事情:
import matplotlib.pyplot as plt
import numpy as np
from scipy.optimize import leastsq
def data_func( x, x0, a, bc, off, p1, p2):
"""
fit function that uses modulus to get periodicity
two linear functions are combines piecewise by boxing them
using the heaviside function with the periodic input
over all slope is added.
Note that the "decay" part maybe positive with this solution.
"""
P1 = abs(p1)
P2 = abs(p2)
X = x - x0
P= P1 + P2
mod = X % P
y0 = a * P1
beta = y0 * P / P2
slope = y0 / P2
box1 = np.heaviside( +np.abs( ( X - P1 / 2. ) % P - 0.5 * P ) - 0.5 * P2, .5 )
box2 = np.heaviside( -np.abs( ( X - P1 / 2. ) % P - 0.5 * P ) + 0.5 * P2, .5 )
out = a * mod * box1
out += (beta - slope * mod )* box2
out += off + bc * X
return out
def residuals( params, xl ,yl ):
x0, a, bc, off, p1, p2 = params
diff = np.fromiter( ( y - data_func( x, x0, a, bc, off, p1, p2 ) for x, y in zip( xl, yl ) ), np.float )
return diff
theOff = 0.7
theP1= 1.8869
theP2 = 5.21163
theP = theP1 + theP2
xdata = np.linspace(-1, 26, 51 )
xdata = np.fromiter( ( x for x in xdata if (x-theOff)%theP <= theP1 ),np.float )
ydata = np.fromiter( ( data_func( x, theOff, .6, .1, 17, theP1, theP2) for x in xdata ),np.float )
tl = np.linspace(-1, 26, 150 )
yl = np.fromiter( ( data_func( x, theOff, .6, .1, 17, theP1, theP2) for x in tl ),np.float )
guess= [0, 0.55, .1, 16 , 2, 5 ]
sol, err = leastsq( residuals, guess, args = ( xdata, ydata ) )
print sol
### getting the real slopes out of the data
s1 = sol[1]+ sol[2]
s2 = - sol[1] * sol[4] / sol[5] + sol[2]
print "real slope1 = {}".format( s1 )
print "real slope2 = {}".format( s2 )
fit = np.fromiter( ( data_func( x, *sol ) for x in tl ),np.float )
fig = plt.figure()
ax = fig.add_subplot( 1, 1, 1 )
### original data
ax.plot( tl, yl, ls='--')
ax.plot( xdata, ydata, ls='', marker='+')
### fit
ax.plot( tl, fit )
### check the slopes
short = np.linspace(0, 3, 3)
ax.plot( short, [ 17 + s1 * s for s in short ] )
short = np.linspace(3, 10, 3)
ax.plot( short, [ 18 + s2 * s for s in short ] )
ax.grid()
plt.show()
这使:
>> [ 0.39352332 0.59149625 0.10850375 16.78546632 1.85009228 5.35049099]
>> real slope1 = 0.7
>> real slope2 = -0.0960237685357
和
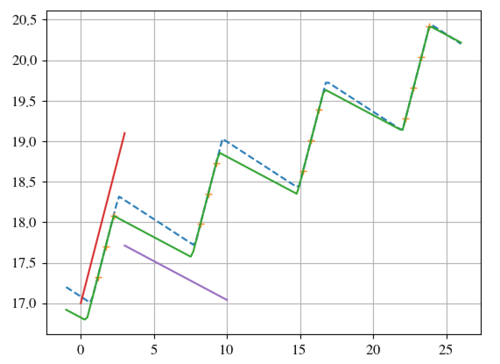
自然地,间隙中缺乏信息会导致那里的斜率相当糟糕。结果,在周期性中存在相应的误差。如果知道这一点,那么准确度当然会提高。
您需要对起始参数进行合理的猜测!
TA贡献1906条经验 获得超10个赞
当假设所有数据都具有相同的斜率m并且所有“衰减”斜率时,那将是我的 AnsatzD
import matplotlib.pyplot as plt ### only for plotting; not important for the OP
import numpy as np ### for easy data manipulation
from scipy.optimize import leastsq ### one of many possibilities for optimization
def generic_data( m, D, n ): ### just generating data; not important for OP
alpha0 = 0
timel = [ 0 ] ### to avoid errer, remove at the end
dl = list()
for gaps in range( n + 1 ):
for pnts in range( 3 + np.random.randint( 2 ) ):
timel.append ( timel[-1] + 0.5 + np.random.rand() )
dl.append( m * timel[ -1 ] + alpha0 )
###now the gap
dt = 2. + 2 * np.random.rand()
timel.append ( timel[-1] + dt )
dl.append( dl[-1] - D * dt )
alpha0 = dl[-1] - m * timel[-1]
del timel[0]
### remove jump of last gap
del timel[-1]
del dl[-1]
dl = np.fromiter( ( y + np.random.normal( scale=0.1 ) for y in dl ), np.float )
return np.array( timel ), dl
def split_into_blocks( tl, dl ):
"""
identifying the data blocks by detecting positions of negative slope.
first subtract a shifted version of the data from itself
find the negatives and get their positions
get sub-lists based on this positins
"""
mask = np.where( dl[1::] - dl[:-1:] < 0, 1, 0 )
where = np.argwhere( mask )
where = where.reshape( 1, len( where ) )[0]
where = np.append( where, len( dl ) - 1 )
where = np.insert( where, 0, -1 )
tll = list()
dll = list()
for s, e in zip( where[ :-1:], where[ 1:: ] ):
dll.append( dl[ s + 1 : e + 1 ] )
tll.append( tl[ s + 1 : e + 1 ] )
return np.array( tll ), np.array( dll )
def residuals( params, tblocks, dblocks ):
"""
typical residual function to be called by leastsq
"""
### split parameters
nob = len( tblocks )
m = params[0] ### all data with same slope
D = params[1] ### all decay with same slope
alphal = params[2:2+nob] ### off sets differ
betal = params[-nob+1::]
out = list()
### evaluate diefference between data and test function
for i, (tl, dl) in enumerate( zip(tblocks, dblocks ) ):
diff = [ d - ( m * t + alphal[i] ) for t, d in zip( tl, dl ) ]
out= out + diff
### evaluate differences for the gapfunction; this could be done
### completely independent, but I do it here to have all in one shot.
for j in range( nob -1 ):
out.append( dblocks[ j][-1] - ( betal[j] + D * tblocks[j][-1] ) ) ###left point gap
out.append( dblocks[ j+1][0] - ( betal[j] + D * tblocks[j+1][0] ) ) ###right point gap
return out
### create generic data
tl, dl = generic_data( 1.3, .3, 3 )
tll, dll = split_into_blocks( tl, dl )
### and fit
nob = len(dll)
m0 = +1.0
D0 = -0.1
guess = [m0, D0 ]+ nob * [-3] + ( nob - 1 ) * [ +4 ]
sol, err = leastsq( residuals, x0=guess, args=( tll, dll ) )
mf = sol[0]
Df = sol[1]
print mf, Df
alphal = sol[2:2+nob]
betal = sol[-nob+1::]
### ... and plot
fig = plt.figure()
ax = fig.add_subplot( 1, 1, 1 )
###original data
ax.plot( tl, dl, ls='', marker='o')
###identify blocks
for a,b in zip( tll, dll ):
ax.plot( a, b, ls='', marker='x')
###fit results
for j in range(nob):
tloc = np.linspace( tll[j][0] - 0.3, tll[j][-1] + 0.3 , 3 )
ax.plot( tloc, [ mf * t + alphal[j] for t in tloc ] )
for j in range(nob - 1):
tloc = np.linspace( tll[j][-1] - 0.3, tll[j+1][0] + 0.3 , 3 )
ax.plot( tloc, [ Df * t +betal[j] for t in tloc ] )
plt.show()
这导致
>> 1.2864142170851447 -0.2818180721270913
和
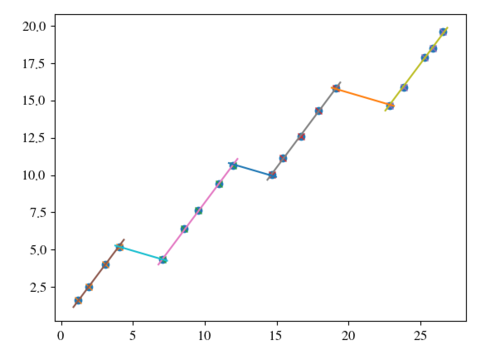
然而,该模型可能要求衰减线不与数据范围内的数据线相交。这使得额外的摆弄成为必要,因为我们需要检查某种类型的边界。另一方面,可以只拟合数据并寻找具有满足前面提到的边界的最小可能斜率的衰减曲线。因此,在这种情况下,我会D从残差中删除拟合部分,然后再计算它。
添加回答
举报
